GREASE THOSE WHEELS ( BIOLOGY )
#3 GENETIC CONTROL
People say that genetics is a unit that simply blasts off the thought of an easy biology course. I, myself, have spoken to many-a-students who have dropped bio' for it and those who run away from the very thought of base-pairing, DNA and RNA.
Therefore, this post is dedicated to all those who think genetics is crap!!!
Lets start with the structure of DNA and RNA.
DNA, short for deoxyribonucleic acid is the longest molecule in your body, so long that when all the DNA in your body is put together it could go to and come back from the sun nearly 102 times!!!
Lets start with its basic structure...
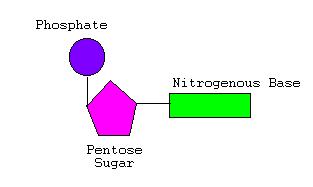
DNA is made up of basically these three things...
- Deoxyribose Sugar (A type of pentose aka 5-Carbon sugar, variant of ribose as it lacks O at 2' Carbon)
- Phosphates. x3 but the two break-away when phospho-diester bond forms.
- Nitrogenous Bases which include...(they are all flat structures because of being rings and thus are easily stackable in the DNA molecule which compacts the size and also strengthens the molecule)
- Purines (dual-ring structure)
- Adenine
- Guanine
- Pyrimidines (single-ring structure consisting of six sides)
- Thymine
- Cytosine
All of the above mentioned components, together build the backbone of the DNA molecule. They form nucleotides (see left) which get linked together to form a polynucleotide.
Now let us see the DNA structure in more detail...
First of all there is a single strand...
- Then the polynucleotide chain is formed which contains half the amount of sugar, phosphate groups and bases than in the final DNA molecule.
- polynucleotide arrangement is achieved by linking nucleotides together with the aid of phosphate groups. The phosphate attaches either to the 5' of one sugar and the 3' to the adjacent one or vice versa (in a phospho-diester bond that is). The former is the case with the above illustrated nucleotide and therefore the orientation of the formed polynucleotide will be from 5'-3' and the other DNA strand will be in an anti-parallel orientation of 3'-5', this kind of arrangement helps in the hydrophilic sugar and phosphate groups facing outwards and the hydrophobic bases to face inwards thus resulting in the stability of the DNA molecule by twisting it around as the sides pull the molecules in the opposite direction.
Then its mate shows up...
- both the polynucleotides join in an anti-parallel arrangement with complimentary base pairing. The hydrogen bonds between adenine-thymine and guanine-cytosine link the strands together.
Anti-parallel structure of DNA. The red/white molecule is the OH on the 3' of deoxyribose. While the orange\red molecule if the Phosphate one, attached to the 5' of ribose.
Complimentary Base arrangement in DNA. Cytosine-Guanine (left) and Adenine-Thymine (right). White, broken lines show hydrogen bond.
Some important points in a DNA's arrangement are...
- Hydrogen bond, although relatively weak when compared to covalent bonds, become a strong force when occurring between millions of base pairs. This confers stability on the vital DNA molecule and helps it stay together. Hydrogen bonds also aid in the formation of the dual-helix structure of DNA. Their weak nature also helps in the easy unzipping of the two strands of DNA when required.
- Accurate base-pairing is essential in a DNA molecule as an alteration in these would result in mutations that several affect a living organism. Each combination of nucleotides is a gene codes for a specific polypeptide and thus changes in these would result in a wrong polypeptide being encoded.
Now lets get to the RNA...short for ribonucleic acid; it is a single strand molecule that never has thymine but has uracil instead. It also contains ribose sugar instead of deoxyribose sugar in its nucleotides. There are three main types of RNA in our body.
- Messenger RNA (mRNA) is formed in the nucleus as a mirror image of a strand of DNA, except that thymine is replaced by uracil. It is a alpha-helical in structure and acts as a template for protein synthesis.
- Transfer RNA (tRNA) is also formed in nucleus from about 80 nucleotides and has a clover-leaf shape. Its main role is to fetch amino acids for protein synthesis and has specificity for them thus about 20 types of tRNAs exist for the twenty amino acids in nature.
- Ribosomal RNA (rRNA) is the central component of our protein manufacturing factories, i.e, ribosomes. It is the largest of the RNAs and can also exist in dual-helical form. Its main function is the decoding of the mRNA molecule and interaction with the tRNA the results in a polypeptide being formed.
Over with the structures now let us get to some factory work!!!
1st Process....REPLICATION....
The Crew....
------------------------------------------------------------------------------------------------------------------------
2nd Process...TRANSCRIPTION....
The Crew....
------------------------------------------------------------------------------------------------------------------------
------------------------------------------------------------------------------------------------------------------------
------------------------------------------------------------------------------------------------------------------------
1st Process....REPLICATION....
The Crew....
- Initiator Proteins: provide origin for replication by forming a replication bubble or opening.
- Helicase: unzips or opens up the DNA strand.
- SSB: or single-strand-binding proteins temporarily bond with the DNA strands to prevent them from recombining.
- Gyrase: prevents knotting in the DNA open. molecule
- Primase: lays down primers so that replication can initiate.
- DNA Polymerase: it is the main player in DNA replication and is an enzyme complex that carries out the following functions...
- Adding of nucleotides.
- Replacing RNA Primers with DNA.
- Proofreading.
- Ligase: seals the gaps on the lagging strand.
The process...
- Initiator protein scoots along the DNA molecule until it comes across a specific base combination. it stops there and creates a small opening.
- Helicase then finds that opening and unzips the DNA molecule into two strands by breaking the hydrogen bonds, forming a replication fork, or a horizontal Y in the DNA molecule.
- SSB proteins then bind with the separated strands so that they don't recombine again.
- Gyrase prevents the knotting over of the coiled DNA molecule as in a separated state, it has a real tendency to disrupt our genetic code.
- As for a new strand to begin some foundation should be laid to construct over, i.e, a reactive OH tail at 3' of the deoxyribose sugar should be present so that a polynucleotide can form from it. So as the new strand is completely naked i.e has nothing to support the formation of a new strand, primers (RNA nucleotide) is laid by the enzyme RNA primase to get the process rolling. However there is a complex situation involved. DNA Polymerase can only add deoxyribonucleotides (a fancy way to say, DNA's nucelotides) in the 5'-3' direction or in other words only to a free 3' OH tail; this results in a leading and lagging strand. The leading strand being where DNA polymerase keeps on adding ribonucleotides in the 5'-3' direction continuously after an initial primer is placed. The lagging strand being the one where addition of ribonucleotides occurs in breaks called, Okazaki fragments, which are around 200-300 nucleotides long and polymerase stops when it encounters 5' of another fragment. It is a difficult concept, I know, therefore lets have a look at the diagram below.
- Ligase fills the gaps between the fragments. As DNA polymerase replaces the RNA primers with DNA.
- Finally proof-reading is done by DNA Polymerase in 3'-5' direction.
2nd Process...TRANSCRIPTION....
The Crew....
- RNA Polymerase: does the same work as DNA Polymerase in the earlier section but has only two fundamentally different functions than the DNA Polymerase...
- Uracil is used instead of thymine.
- Usage of ribonucleotide instead of deoxyribonucleotide in polynucleotide assembly.
- Holoenzyme: is a group of enzymes that look up the gene to be transcribed for the RNA Polymerase.
The Process....
- Holoenzyme finds the promoter sequence for RNA Polymerase. It is usually known as the TATA Box and is on the non-template strand (the one to be transcribed); the promoter tells RNA Polymerase that how many bases away is the base sequence of the gene to be encoded.
- After Holoenzyme has found the promoter gene; it signals RNA Polymerase to come over and begin the task of transcription.
- RNA Polymerase first melts the hydrogen bonds between the two strands so that the region to transcribe can be viewed.
- When the template strand (the one that is not transcribed but serves as a template to form the non-template strand) is exposed. RNA Polymerase starts laying down ribonucleotides which have bases complimentary to the template strand without the need of a primer.
- The elongation of the mRNA strand proceeds in the 5'-3' direction until the terminator sequence is encountered by the DNA Polymerase upon which it transcribes the terminator sequence and halts transcription.
- It detaches itself from the template strand.
- A 5' Cap of Guanine is added to the mRNA molecule with some methyl groups that prevent decomposition and deem the mRNA molecule ready for translation.
- A 3' Tail of Adenine is attached at the end of the mRNA molecule, again to prevent decomposition form nucleases as mRNA is temporary so nucleases start destroying it the moment it steps into the cytoplasm so the addition of tails means that the message stays long enough to be translated by ribosomes.
- Intorns (non-encoding)are removed from the mRNA molecule so that only Exons (encoding) remain.
3rd Process...TRANSLATION....
The crew...
- mRNA: serves as the template for translation.
- Amino Acids: they are linked together to form a protein.
- tRNA: provides courier service during the process by fetching the amino-acids required.
- Ribosomes: provide the frame-work for the occurring of the process.
The process...
- mRNA moves out into the cytoplasm.
- tRNA have their complimentary amino acid hooked to the acceptor / 3' arm by aminoacyl synthetases who recognize the anti-codon on the tRNA molecule.
- The two subunits of Ribosomes come together, the smaller subunit binds to the 5' end of the mRNA molecule.
- Codon (group of three bases) on the mRNA molecule attract their complimentary anticodons on the tRNA molecule. The ribosome unit that is travelling along the mRNA strand allows the incoming complimentary tRNA molecules to fit in; two at a time, when the two tRNA molecules are held in close proximity, then the amino acids on their 3' arms combine to form a peptide bond and thus a polypeptide forms as the chain grows.
- Many ribosomes work on the same tRNA molecule to give many copies of the protein transcribed on the mRNA molecules.
- When the terminator sequence is reached by the ribosome, then translation stops as there is no anticodon for it.
Structure of tRNA
...CODON TABLE...
One of the most common anomalies associated with wrong base sequence, is none other than the sickle-cell anaemia. Lets journey to the core of this disease.
Name: Sickle-Cell Anaemia
Affected body-parts: Blood Tissue \ RBCs
What it does?: This genetically inherited disease alters the shape of the RBC; from flattened biconcave disks to crescent or sickle shaped. This reduces the amount of oxygen they can carry and also makes them 50-times less soluble in blood upon deoxygenation which can result in the cells being precipitated.
Symptoms: Body Ache, Lethargy, Pale Skin.
Cause: Normally our Hemoglobin (HbA) has four polypeptide chains (alpha, beta, delta and gamma) which combine with 4 haem groups. But in sickle cell patients, due to wrong base grouping, valine (see above, its an amino acid) replaces glutamine (see above) at the 6th position in the beta-polypeptide chain, such small a change can lead to such drastic effects. But this disease shows in up in only homozygous form, i.e, when both the genes you got from your parents code for sickle cell RBCs. In hetrozygous form, you can only have a sickle-cell trait but, luckily, not the disease!!
Prevention: No preventive measure known.
Medication: Pain-relieving medicines, Hydrea, Droxea, Blood Transfusions, Supplemental Oxygen, Bone-Marrow transplant, Nicosan.
Special Fact: Reduced susceptibility to the malarial parasite.









No comments:
Post a Comment